Edge/Node Weighted Watersheds¶
Compare edge weighted watersheds and node weighted on a grid graph.
sphinx_gallery_thumbnail_number = 5
from __future__ import print_function
import nifty.graph
import skimage.data
import skimage.segmentation
import vigra
import matplotlib
import pylab
import numpy
# increase default figure size
a,b = pylab.rcParams['figure.figsize']
pylab.rcParams['figure.figsize'] = 2.0*a, 2.0*b
load some image
img = skimage.data.astronaut().astype('float32')
shape = img.shape[0:2]
#plot the image
pylab.imshow(img/255)
pylab.show()

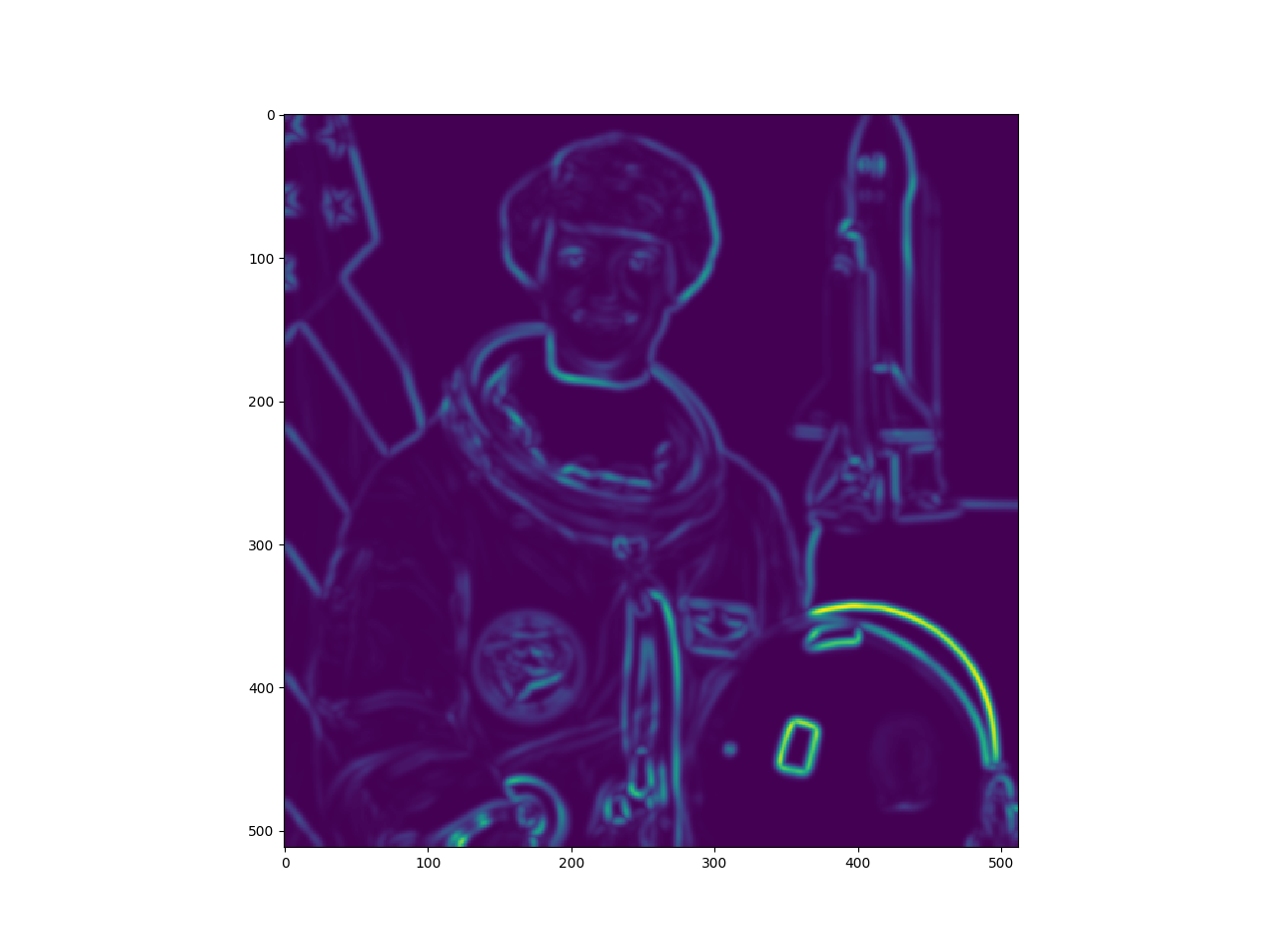
get some edge indicator
taggedImg = vigra.taggedView(img,'xyc')
edgeStrength = vigra.filters.structureTensorEigenvalues(taggedImg, 1.5, 1.9)[:,:,0]
edgeStrength = edgeStrength.squeeze()
edgeStrength = numpy.array(edgeStrength)
pylab.imshow(edgeStrength)
pylab.show()
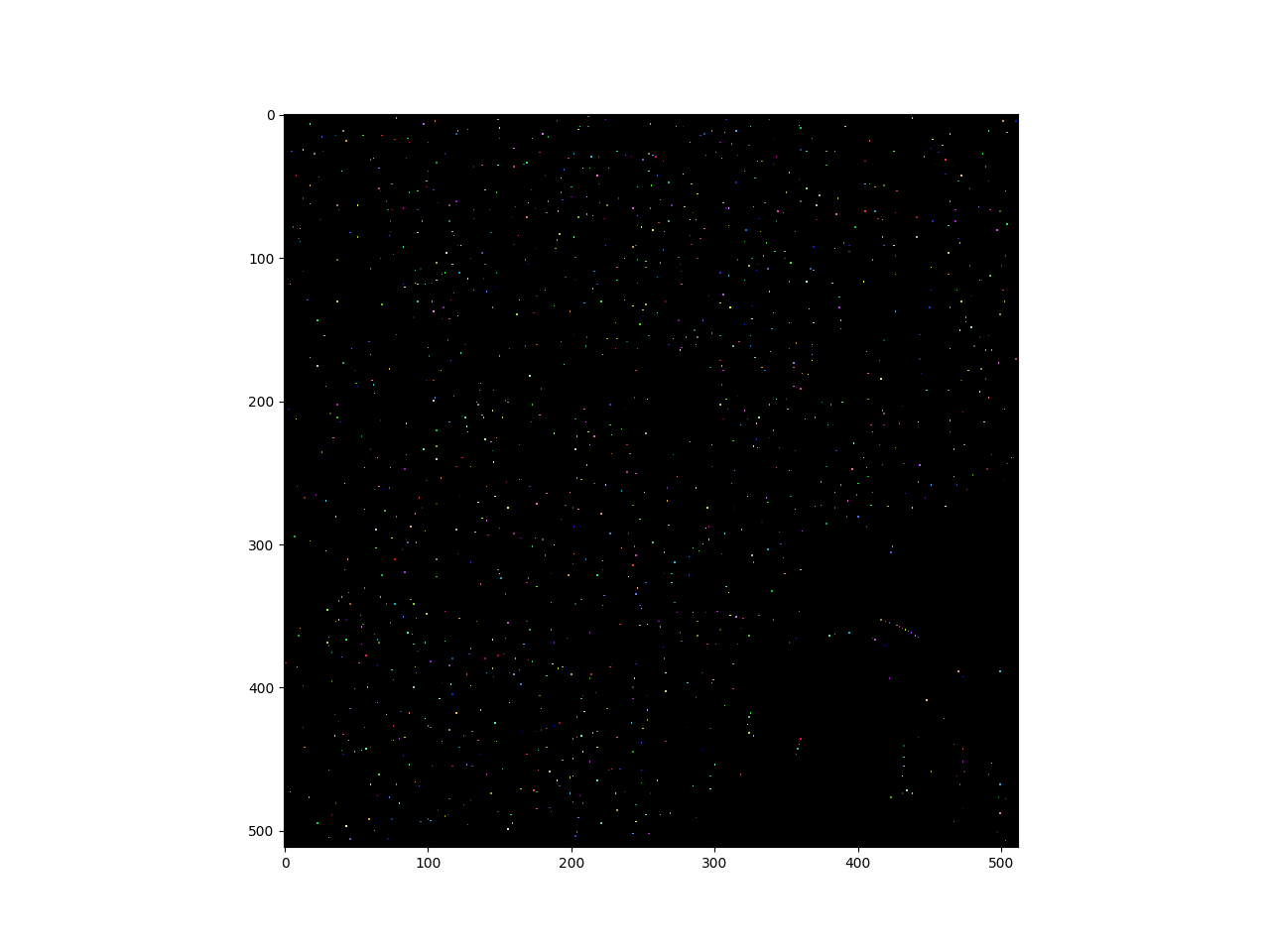
get seeds via local minima
seeds = vigra.analysis.localMinima(edgeStrength)
seeds = vigra.analysis.labelImageWithBackground(seeds)
# plot seeds
cmap = numpy.random.rand ( seeds.max()+1,3)
cmap[0,:] = 0
cmap = matplotlib.colors.ListedColormap ( cmap)
pylab.imshow(seeds, cmap=cmap)
pylab.show()
grid graph
gridGraph = nifty.graph.undirectedGridGraph(shape)
run node weighted watershed algorithm
oversegNodeWeighted = nifty.graph.nodeWeightedWatershedsSegmentation(graph=gridGraph, seeds=seeds.ravel(),
nodeWeights=edgeStrength.ravel())
oversegNodeWeighted = oversegNodeWeighted.reshape(shape)
run edge weighted watershed algorithm
gridGraphEdgeStrength = gridGraph.imageToEdgeMap(edgeStrength, mode='sum')
numpy.random.permutation(gridGraphEdgeStrength)
oversegEdgeWeightedA = nifty.graph.edgeWeightedWatershedsSegmentation(graph=gridGraph, seeds=seeds.ravel(),
edgeWeights=gridGraphEdgeStrength)
oversegEdgeWeightedA = oversegEdgeWeightedA.reshape(shape)
run edge weighted watershed algorithm on interpixel weights. To do so we need to resample the image and compute the edge indicator on the reampled image
interpixelShape = [2*s-1 for s in shape]
imgBig = vigra.sampling.resize(taggedImg, interpixelShape)
edgeStrength = vigra.filters.structureTensorEigenvalues(imgBig, 2*1.5, 2*1.9)[:,:,0]
edgeStrength = edgeStrength.squeeze()
edgeStrength = numpy.array(edgeStrength)
gridGraphEdgeStrength = gridGraph.imageToEdgeMap(edgeStrength, mode='interpixel')
oversegEdgeWeightedB = nifty.graph.edgeWeightedWatershedsSegmentation(
graph=gridGraph,
seeds=seeds.ravel(),
edgeWeights=gridGraphEdgeStrength)
oversegEdgeWeightedB = oversegEdgeWeightedB.reshape(shape)
plot results
f = pylab.figure()
f.add_subplot(2,2, 1)
b_img = skimage.segmentation.mark_boundaries(img/255,
oversegEdgeWeightedA.astype('uint32'), mode='inner', color=(0.1,0.1,0.2))
pylab.imshow(b_img)
pylab.title('Edge Weighted Watershed (sum weights)')
f.add_subplot(2,2, 2)
b_img = skimage.segmentation.mark_boundaries(img/255,
oversegEdgeWeightedB.astype('uint32'), mode='inner', color=(0.1,0.1,0.2))
pylab.imshow(b_img)
pylab.title('Edge Weighted Watershed (interpixel weights)')
f.add_subplot(2,2, 3)
b_img = skimage.segmentation.mark_boundaries(img/255,
oversegNodeWeighted.astype('uint32'), mode='inner', color=(0.1,0.1,0.2))
pylab.imshow(b_img)
pylab.title('Node Weighted Watershed')
pylab.show()

Total running time of the script: ( 0 minutes 2.164 seconds)