Introduction¶
An introduction into the cgp.
# sphinx_gallery_thumbnail_number = 5
from __future__ import print_function, division
import numpy
import scipy
# skimage
import skimage.filters # filters
import skimage.segmentation # Superpixels
import skimage.data # Data
import skimage.color # rgb2Gray
# pylab
import pylab # Plotting
import matplotlib
# increase default figure size
a,b = pylab.rcParams['figure.figsize']
pylab.rcParams['figure.figsize'] = 2.0*a, 2.0*b
import sys; sys.path.append("/home/tbeier/bld/nifty/python/")
# nifty
import nifty
import nifty.cgp
cellNames = ['Junctions','Edges','Regions']
Load image and compute over-segmentation
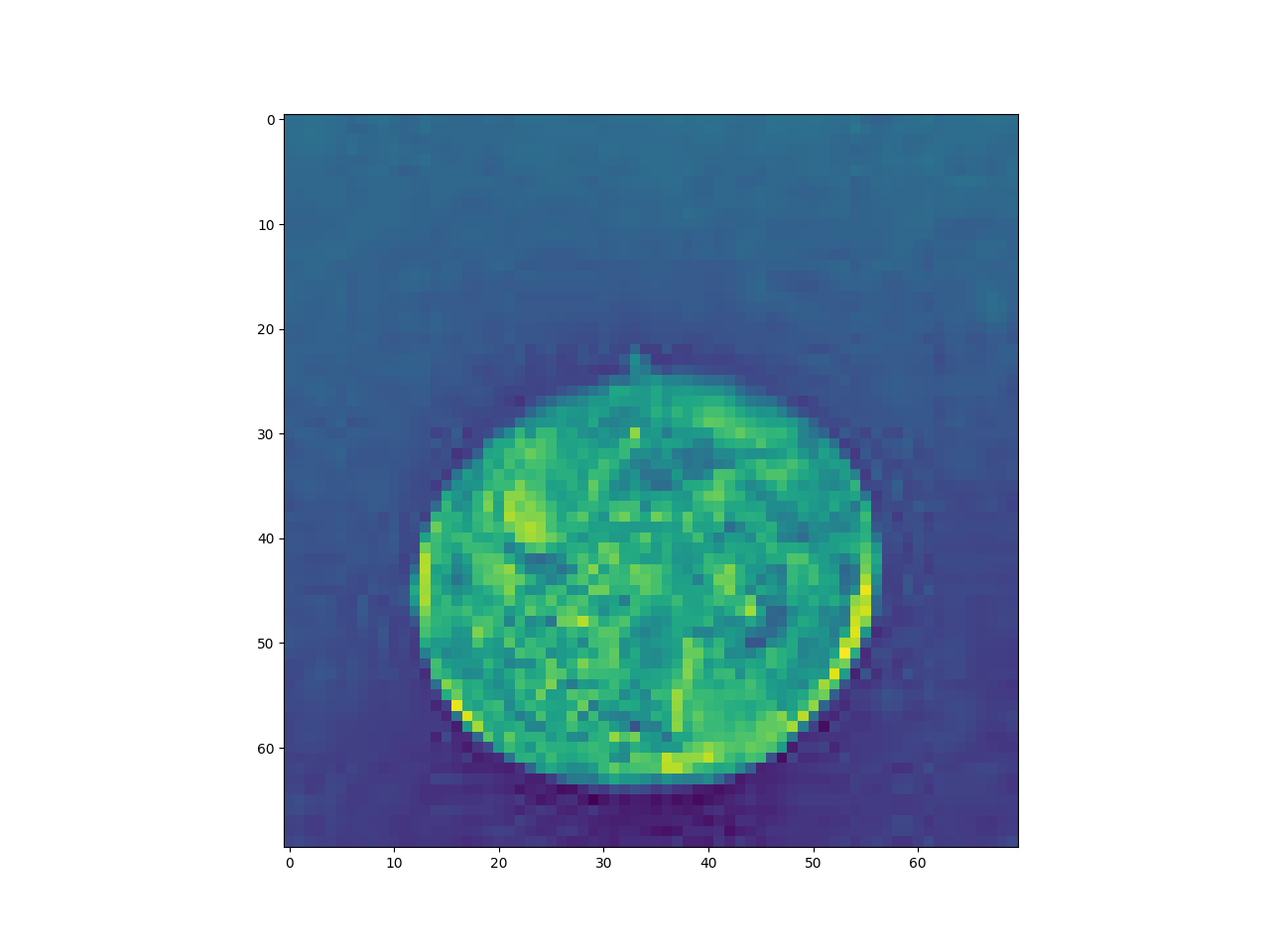
img = skimage.data.coins()[10:80,10:80].astype('float32')/255
pylab.imshow(img)
pylab.show()
# img is a gray value image
# to use it as a rgb img we just
# repeat the gray value
imgRGB = numpy.concatenate([img[...,None]]*3,axis=2)
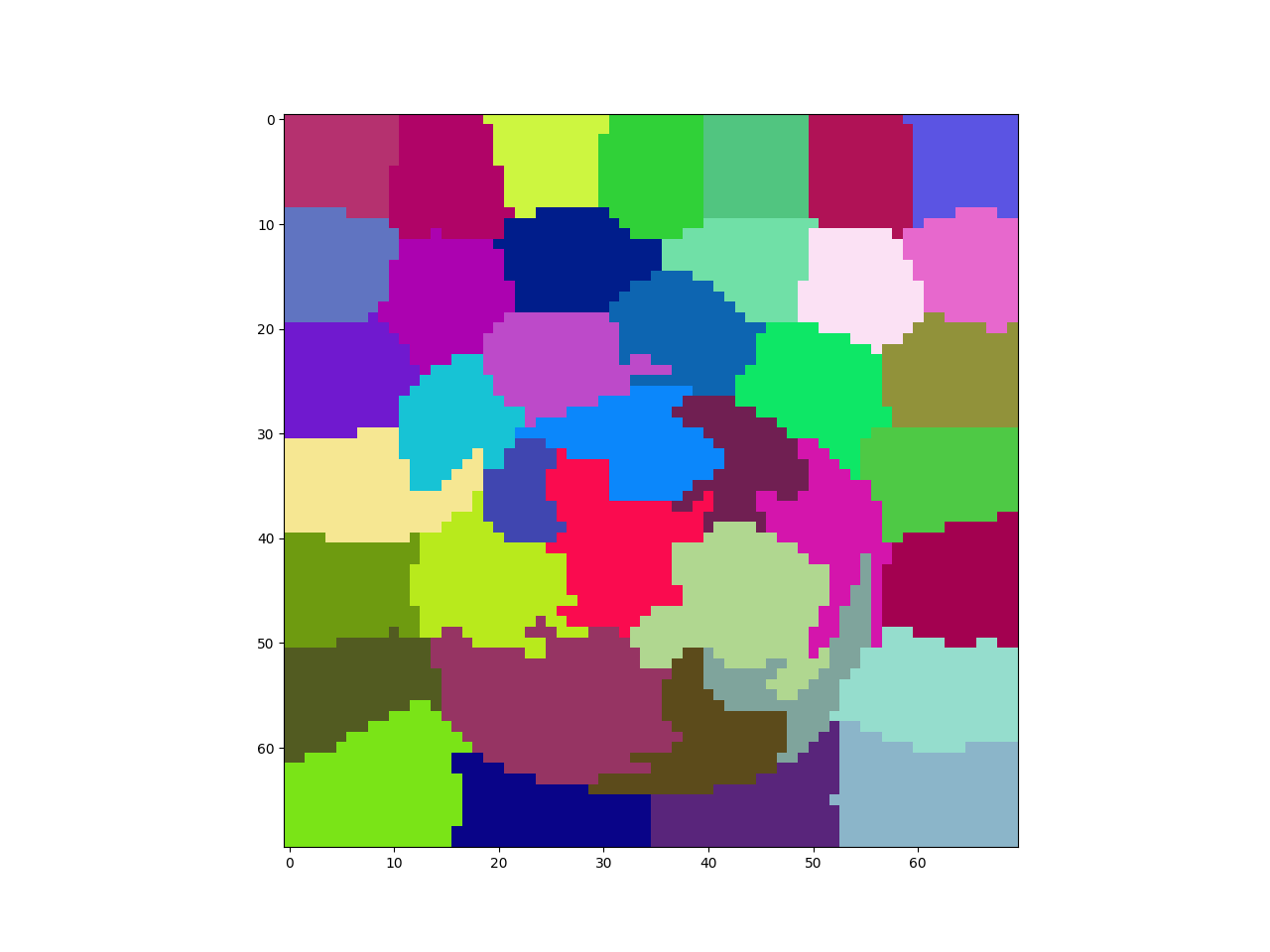
Superpixels
overseg = skimage.segmentation.slic(img, n_segments=50,
compactness=0.04, sigma=1)
# let overseg start from 1
overseg += 1
assert overseg.min() == 1
cmap = numpy.random.rand ( overseg.max()+1,3)
cmap = matplotlib.colors.ListedColormap(cmap)
pylab.imshow(overseg, cmap=cmap)
pylab.show()
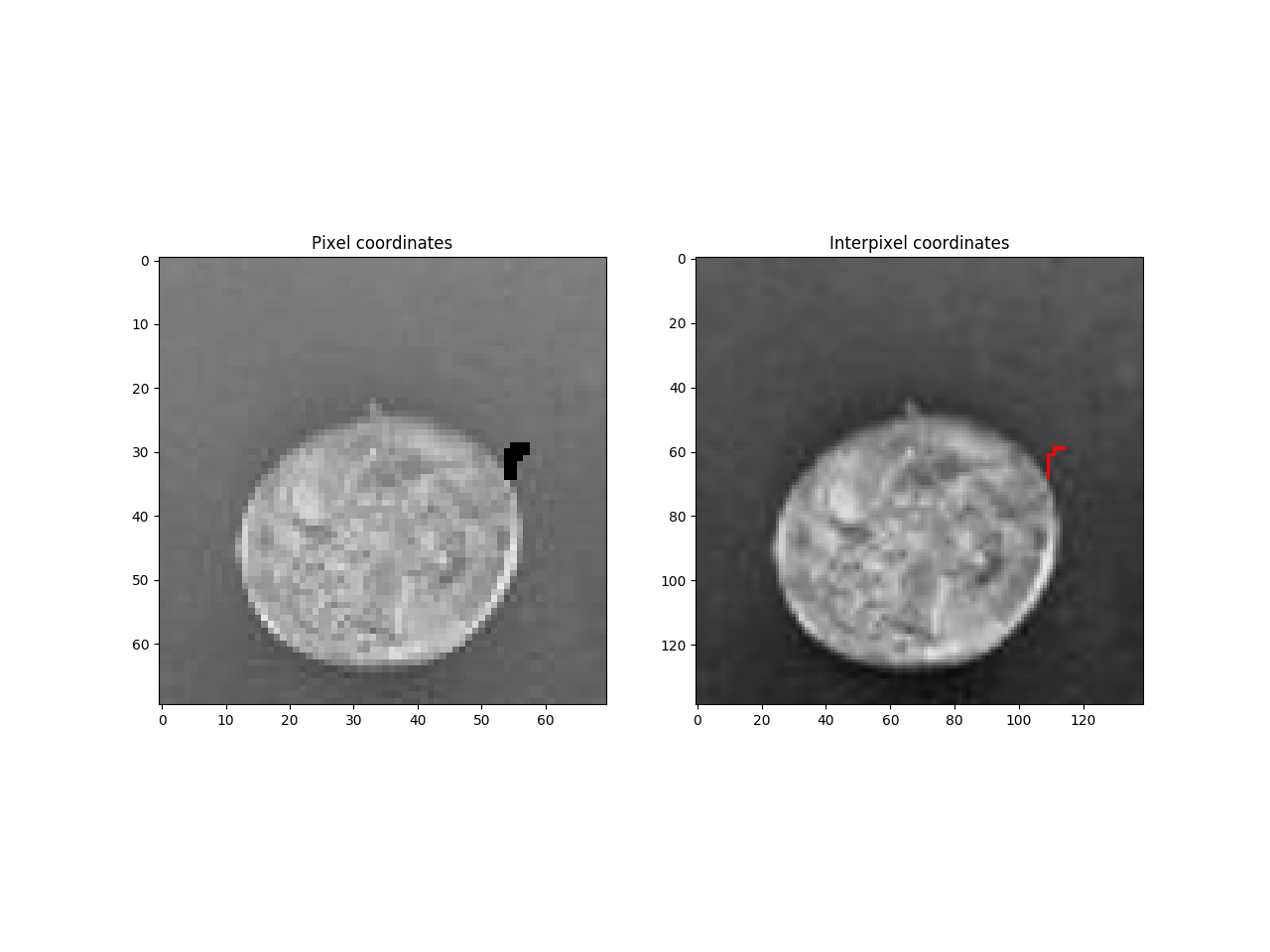
Compute cgp / topological grid
tgrid = nifty.cgp.TopologicalGrid2D(overseg)
Get the geometry of the cell-complex
cellGeometry = tgrid.extractCellsGeometry()
cell0Geomtry = cellGeometry[0]
cell1Geomtry = cellGeometry[1]
cell2Geomtry = cellGeometry[2]
# coordinates of some specific 1 cell
cell1Index = tgrid.numberOfCells[1]//2
coordinates = cell1Geomtry[cell1Index]
# cast coordinates to numpy array
coordinates = numpy.array(coordinates)
# the coordinates are interpixel-coordinates
# to convert them to normal pixels
# we need divide them by 2
# This will lead to non integer coordinates.
# Floor and ceil will lead to integer coordinates
cCoords = numpy.ceil(coordinates/2).astype('uint32')
fCoords = numpy.floor(coordinates/2).astype('uint32')
coords = numpy.concatenate([cCoords,fCoords], axis=0)
# write into image
pixelImg = imgRGB.copy()
pixelImg[coords[:,0], coords[:,1] ,:] = 255,0,0
# we can also use the
# interpixel-coordinates directly
topologicalGridShape = tgrid.topologicalGridShape
print("shape",tgrid.shape, "topologicalGridShape",topologicalGridShape)
interPixelImg = scipy.misc.imresize(imgRGB, topologicalGridShape)
interPixelImg[coordinates[:,0], coordinates[:,1],:] = 255,0,0
f = pylab.figure()
f.add_subplot(1, 2, 1)
pylab.imshow(pixelImg,cmap='gray')
pylab.title('Pixel coordinates')
f.add_subplot(1, 2, 2)
pylab.imshow(interPixelImg,cmap='gray')
pylab.title('Interpixel coordinates')
pylab.show()
Out:
shape [70, 70] topologicalGridShape [139, 139]
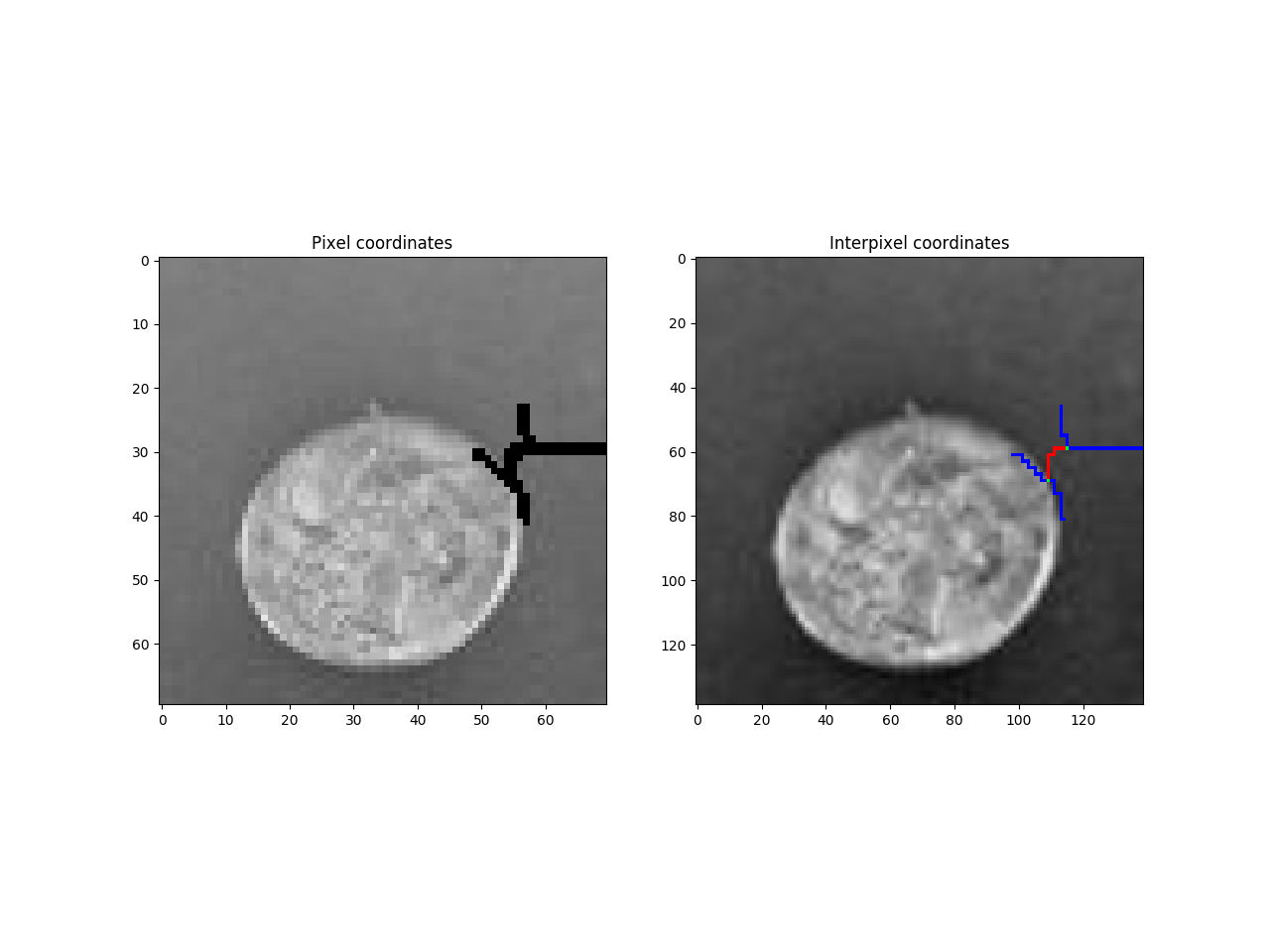
get the bounds and bounded by relations
cellBounds = tgrid.extractCellsBounds()
# this gives you the boundaries (1-cells)
# of a certain junction (0-cell)
cell0Bounds = cellBounds[0]
# this will give us the junctions (0-cells)
# of a certain boundary (1-cell)
cell1BoundedBy = cell0Bounds.reverseMapping()
# get the labels junctions of a particular
# boundary (1-cel)
# Important: Labels start at 1, indexes
# start at 0.
# Bounds/ bounded by returns labels!.
cell0Labels = cell1BoundedBy[cell1Index]
cell0Indices = numpy.array(cell0Labels) - 1
for cell0Index in cell0Indices:
# iterate over the boundaries of this junction
cell1Labels = cell0Bounds[cell0Index]
otherCell1Indices = numpy.array(cell1Labels) - 1
for otherCell1Index in otherCell1Indices:
if otherCell1Index != cell1Index:
coords = numpy.array(cell1Geomtry[otherCell1Index])
interPixelImg[coords[:,0], coords[:,1] ,:] = 0,0,255
cCoords = numpy.ceil(coords/2).astype('uint32')
fCoords = numpy.floor(coords/2).astype('uint32')
coords = numpy.concatenate([cCoords,fCoords], axis=0)
pixelImg[coords[:,0], coords[:,1] ,:] = 0,0,255
# junctions only have a single coordinate
# in supixels coordinates
coord = cell0Geomtry[cell0Index][0]
interPixelImg[coord[0], coord[1], :] = (0,255,0)
# in the image, this are 4 pixels
pixelImg[coord[0]//2, coord[1]//2, :] = (0,255,0)
pixelImg[coord[0]//2, coord[1]//2+1, :] = (0,255,0)
pixelImg[coord[0]//2+1, coord[1]//2, :] = (0,255,0)
pixelImg[coord[0]//2+1, coord[1]//2+1, :] = (0,255,0)
f = pylab.figure()
f.add_subplot(1, 2, 1)
pylab.imshow(pixelImg,cmap='gray')
pylab.title('Pixel coordinates')
f.add_subplot(1, 2, 2)
pylab.imshow(interPixelImg,cmap='gray')
pylab.title('Interpixel coordinates')
pylab.show()
Show all cells labels at once
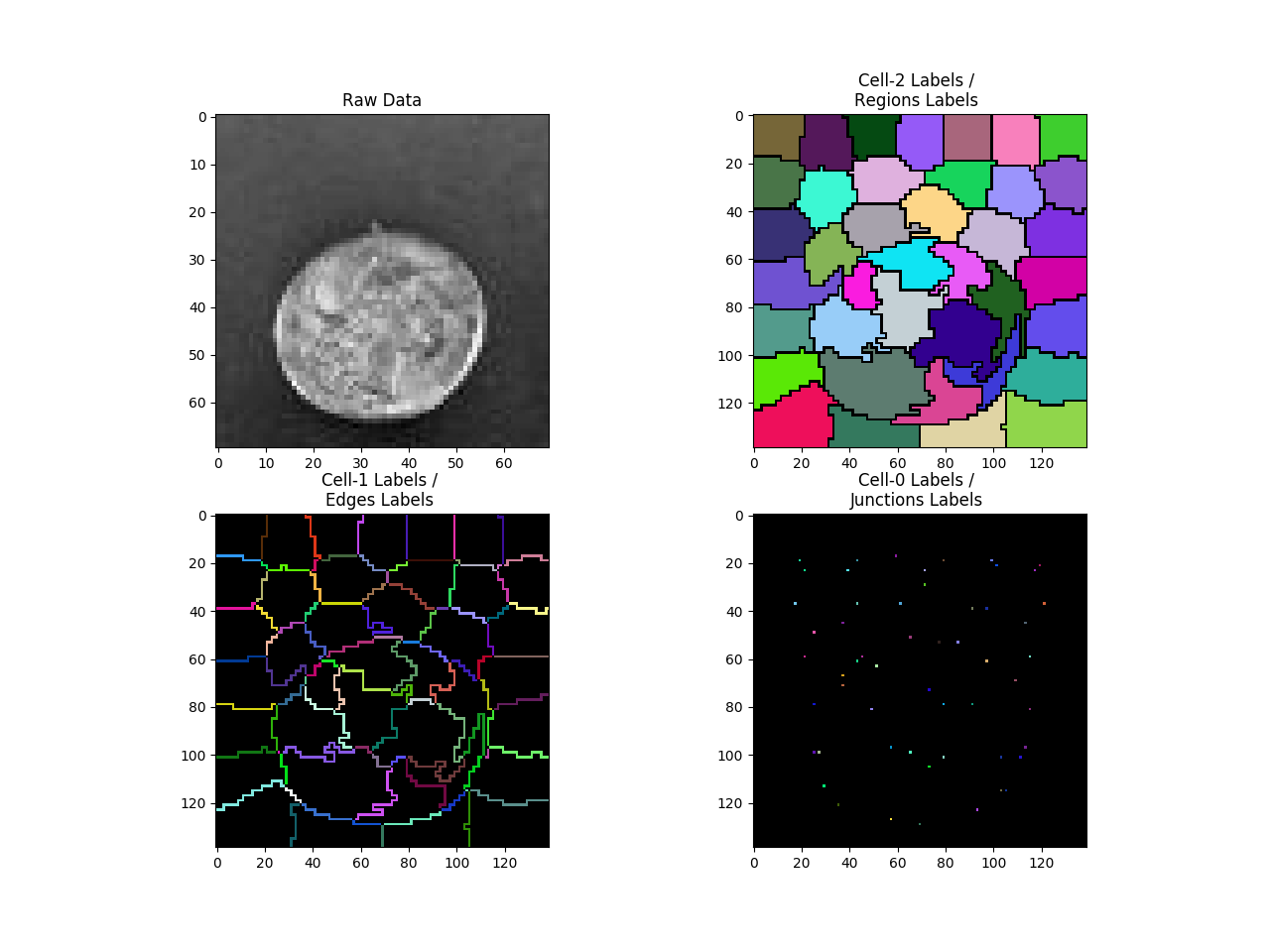
ftgrid = nifty.cgp.FilledTopologicalGrid2D(tgrid)
f = pylab.figure()
f.add_subplot(2, 2, 1)
pylab.imshow(img,cmap='gray')
pylab.title('Raw Data')
cmap = numpy.random.rand ( 100000,3)
cmap[0,:] = 0
cmap = matplotlib.colors.ListedColormap(cmap)
for i,cellType in enumerate((2,1,0)):
showCells = [False,False,False]
showCells[cellType] = True
cellMask = ftgrid.cellMask(showCells)
cellMask[cellMask!=0] -= ftgrid.cellTypeOffset[cellType]
f.add_subplot(2, 2, i+2)
pylab.imshow(cellMask,cmap=cmap)
pylab.title('Cell-%d Labels / \n%s Labels '%(cellType,cellNames[cellType] ) )
pylab.show()
Total running time of the script: ( 0 minutes 0.463 seconds)